Tools
In Vivo Multimodal Imaging
We have a custom built non-invasive, in vivo multimodal imaging system that includes the capacity for optical coherence tomography (OCT), phase-variance OCT (PV-OCT), scanning laser ophthalmoscopy (SLO), and adaptive optics SLO (AO-SLO). We combine these techniques to determine how experimental perturbations affect the retina in the same animal over time.

Viral Vector Design and Packaging
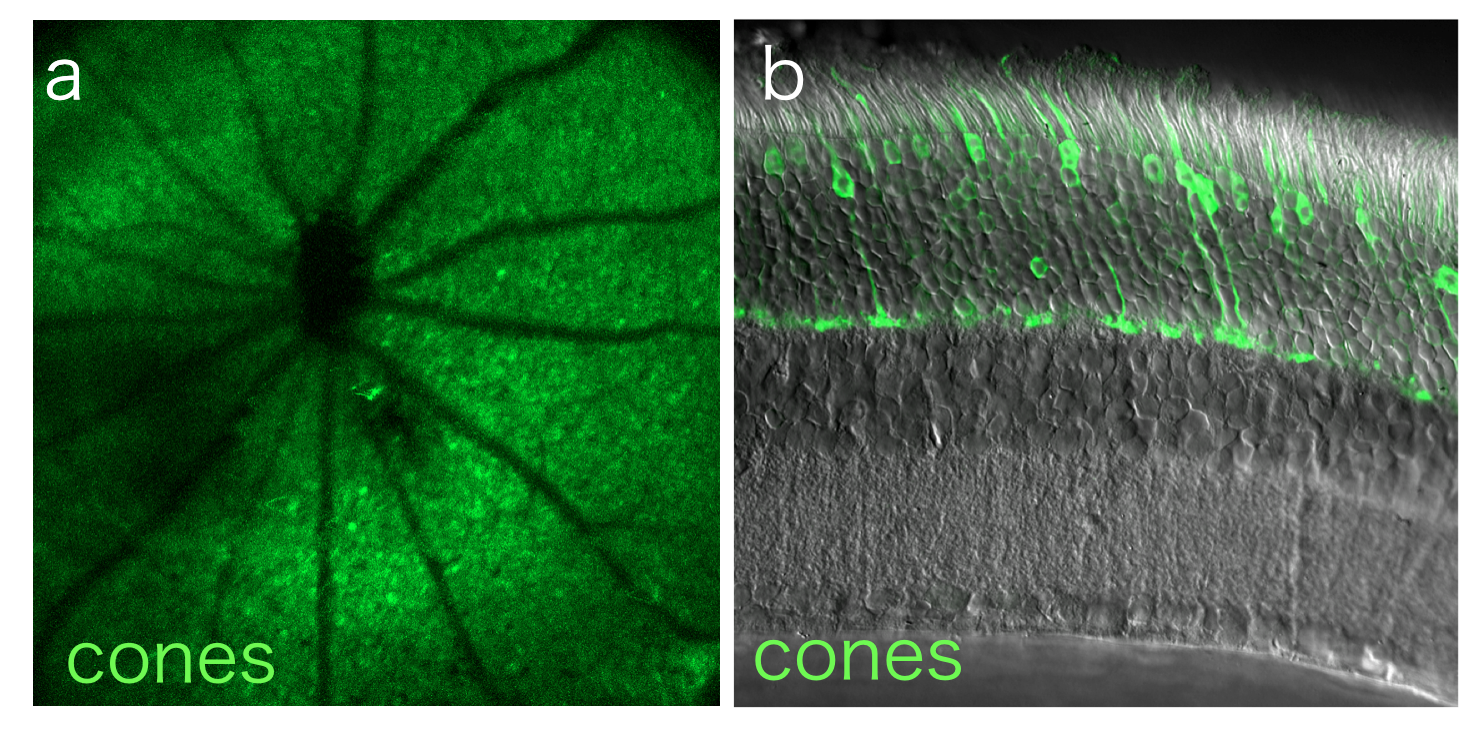
Dr. Burns directs the Molecular Construct and Packaging Core at UC Davis, which designs novel constructs and packages AAV vectors for cell-type specific gene delivery in the eye. We utilize this resource to develop novel AAV7m8-mediated constructs to deliver fluorescent proteins to targeted retinal cell types.
Flow Cytometry
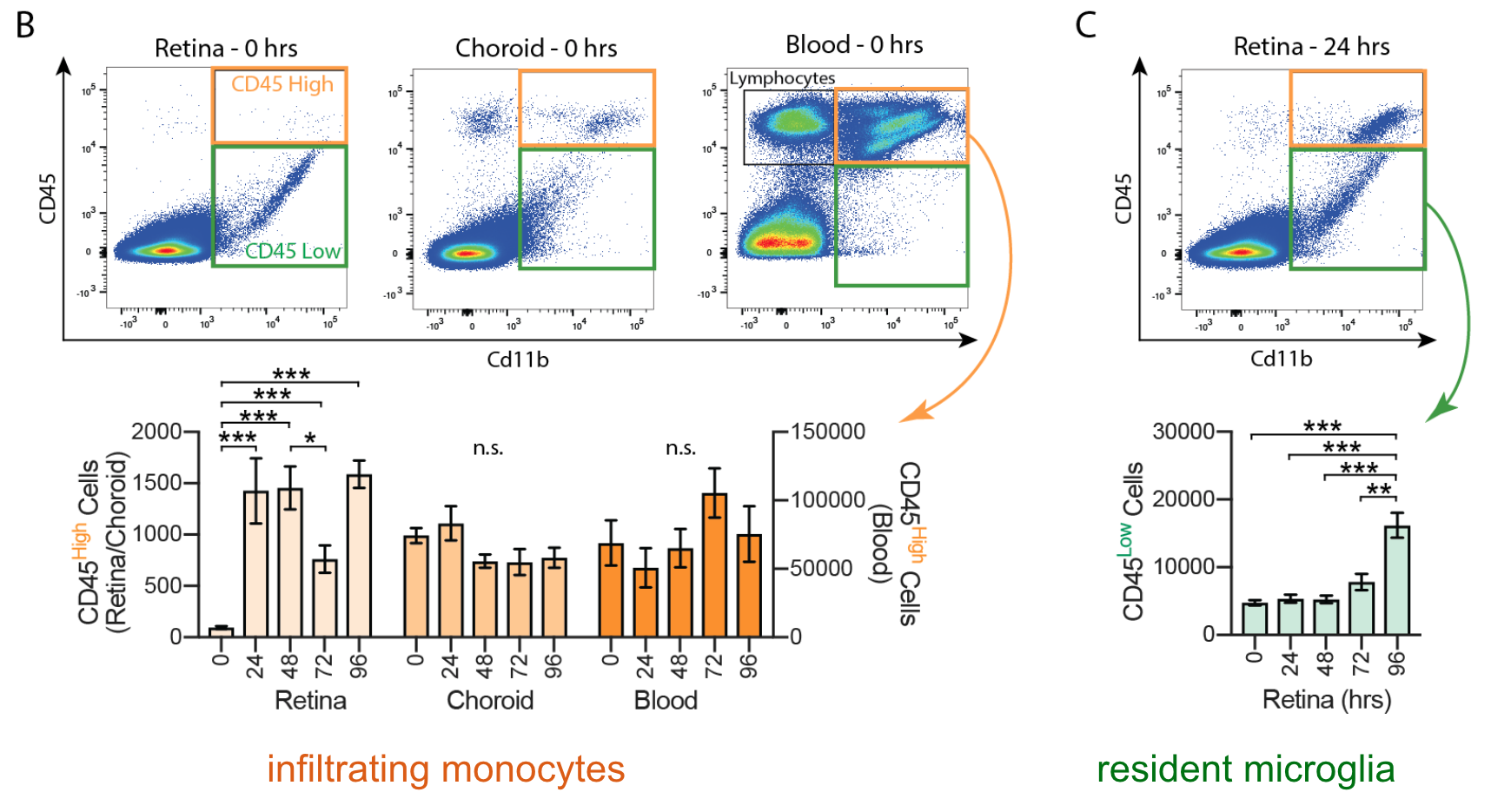
Flow cytometry measures the physical size and fluorescent intensity of a population of dissociated cells stained with fluorescently tagged dyes or antibodies. We use both traditional flow cytometry and fluorescence-activated cell sorting (FACS) to identify and characterize targeted retinal cell populations.
Whole Cell Recording
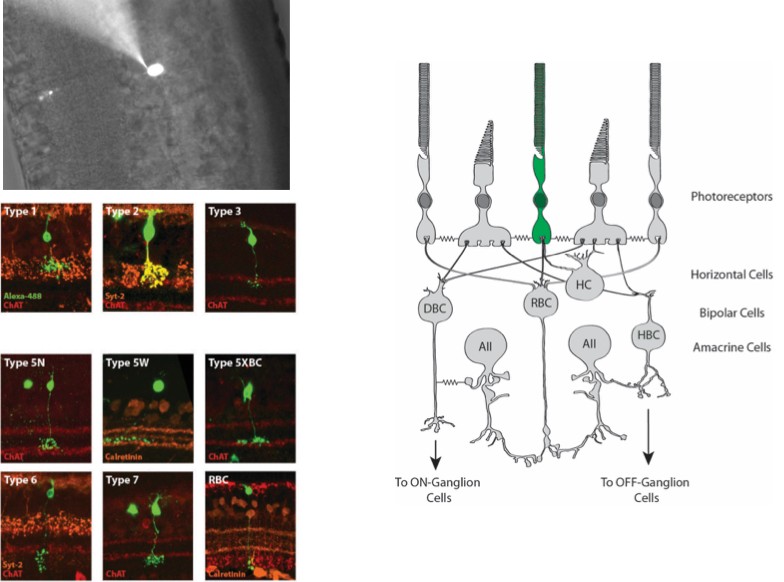
The retina can be embedded in agarose and cut into thick cross sections for whole cell recording. This allows targeting of neuronal or glial cell somas or other cellular compartments for intracellular recording.
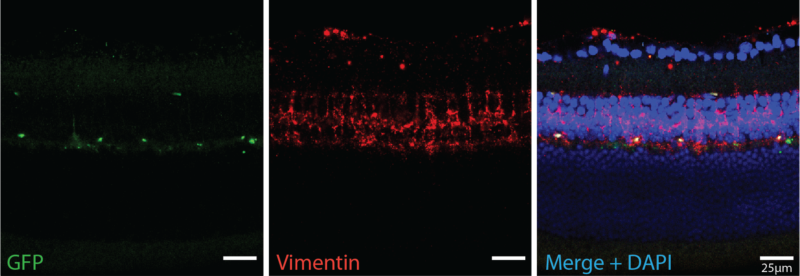
Immunohistochemistry
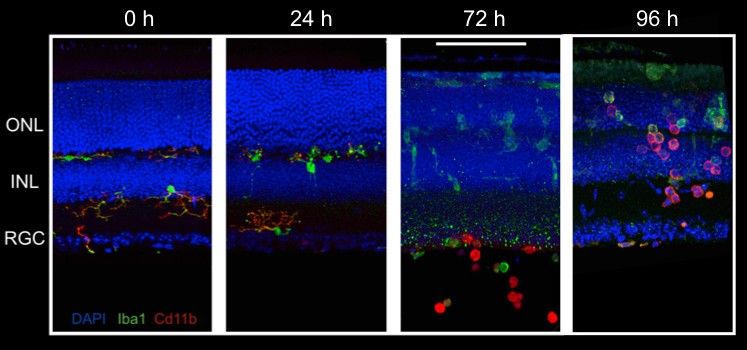
Immunohistochemistry (IHC) allows us to visualize the location of specific proteins by exploiting antibody binding specificity. We use IHC to visual the localization of proteins of interest within the retina to determine how these proteins change over time. We also use proteins as a proxy to identify rental cell types and ask how they respond to experimental perturbation.
Single Cell RNA Sequencing (scRNAseq)
scRNAseq uses next-generation sequencing to determine the transcriptome of gene expression within a single cell. We use scRNAseq, often in combination with FACS, to study the expression profiles of targeted retinal cell populations.
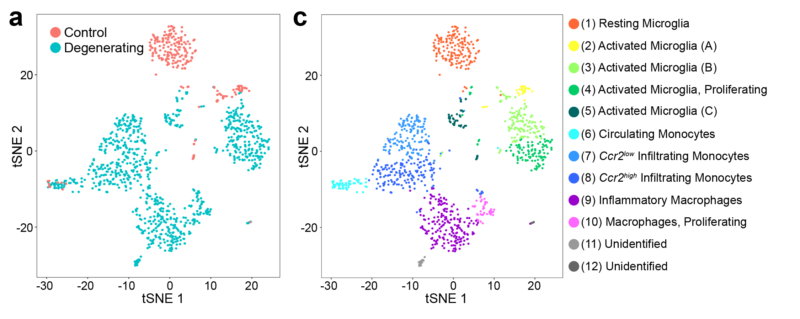
In situ Hybridization
In situ hybridization uses labeled probes to localize complementary RNA sequences. We use this technique to visualize where specific RNA transcripts are located within the retina. We combine in situ hybridization with quantifiable RNA techniques, like scRNAseq and reverse transcription PCR (RT-PCR), to establish RNA transcript location and quantify the amount.
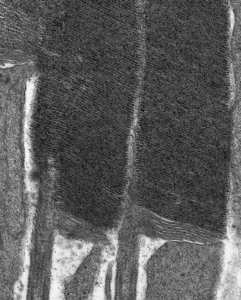
Electron Microscopy
We use transmission electron microscopy to examine the ultrastructure retinal cells and ask how that structure changes over time following various types of experimental manipulation.
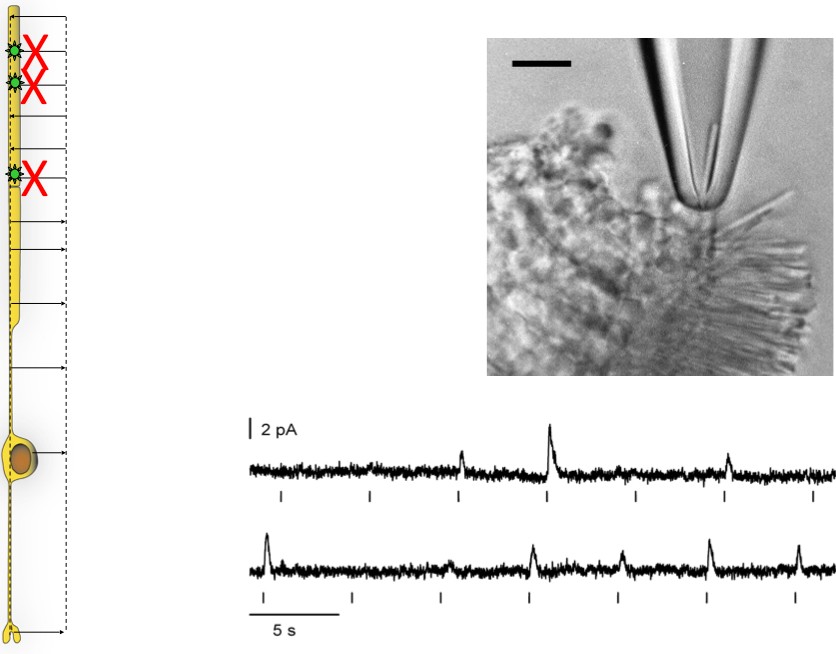
Suction Electrode Recording
Once the retina is pulled away from the pigmented epithelium, the outer segments of photoreceptors are on the surface and can be sucked into an electrode for recording. This approach enables us to measure the amplitude and time course of single photon responses in intact, living rods.
Computational Analysis
We use computational analysis to create detailed models of individual cellular function, as well as multi-cellular interactions within a defined retinal circuit. We use these models to facilitate the interpretation of our anatomical and physiological data.
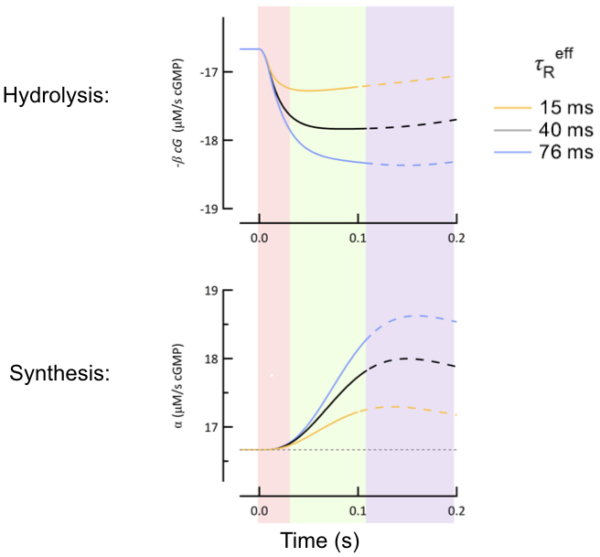
Protein Assays
We use protein assays like Western blots and ELISAs (enzyme-linked immunosorbent assay) to detect and quantify protein levels in retinal tissue.

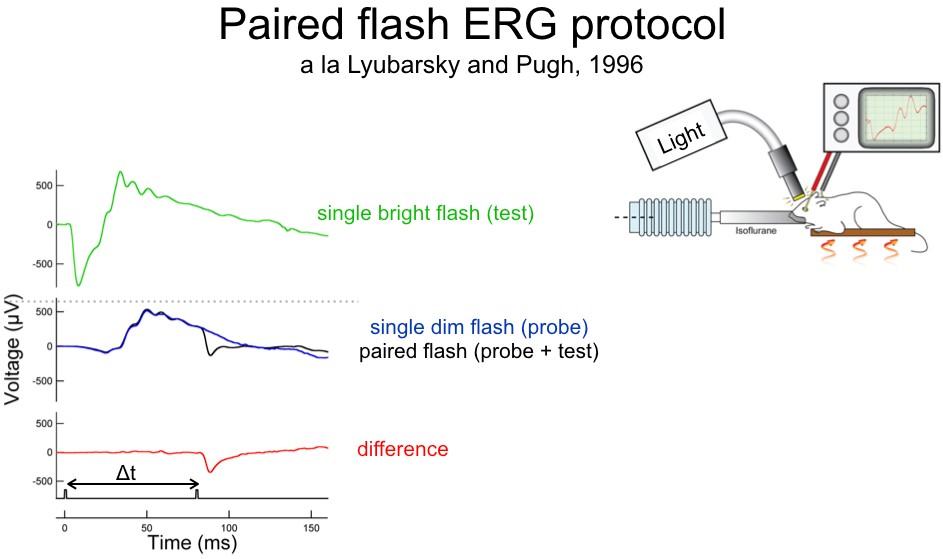
Corneal Electroretinograms
Placing an electrode on the corneal surface of the eye is a non-invasive approach to record overall retinal function. We use ERGs to quantify the response of specific retinal cell types within an intact retinal circuit in order to determine how those responses change under various experimental conditions.
Visually-Guided Behavior
Measuring visually-guided behavior is the only way to determine whether changes in the visual pathway affect an animal’s visual perception. To this end, we utilize a running wheel task to quantify the threshold of visual perception in genetically altered mice.

All image and figure rights reserved. Burns Lab, UC Davis.